Programming Models and Runtime Systems
Have you ever wondered what happens behind the scenes when you write software in a higher-level frameworks like TensorFlow? For example, do you believe that an std function magically sorts your arrays with a single API call? How did we get from explicitly writing quick sort routines to inline function calls? This is where compilers step in and provide wrapper functions that do all the pesky work for you in an optimized fashion while you, as a programmer, get to worry about the more important things.
Ok great! I just explained macros which you already might know. But what happens underneath the quick sort macro. Well, more of the same. You have other helper functions that break down the current helper function. In fact, you can think of them as layers of software. Everything eventually needs to break down to some intermediate representation that your underlying hardware can understand. A good programming model provides a rich set of primitives and interfaces with many layers of indirection.
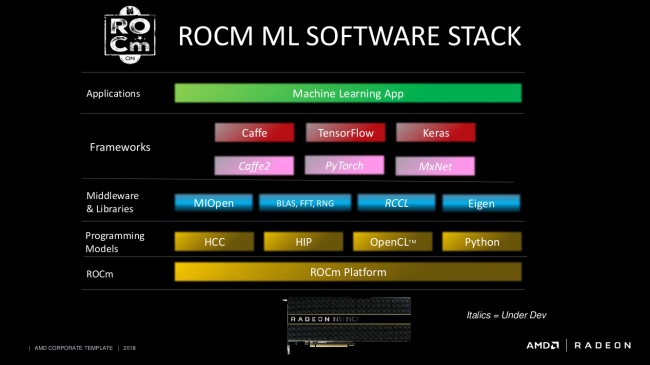
Here is an example of AMD's ROCm stack and all the layers it provides before a piece of your higher-level routine gets executed on their GPUs. Now, this is not going to be the same for all software stacks. Each vendor comes up with its own stack that is highly optimized for its needs.
Let's scope a little deeper into the last layer before your program hits the device. This is the runtime system layer that handles all the necessary scheduling and synchronizations needed to replicate the semantics of your program. After all, a device like a CPU or GPU is nothing but a work-horse that executes the task given in binary. It has no notion of ordering or correctness on its own. The runtime system is in charge of converting complex and interconnected routines written in a higher-level language like C++ into more manageable task packets, and this is done through a process called packetization.
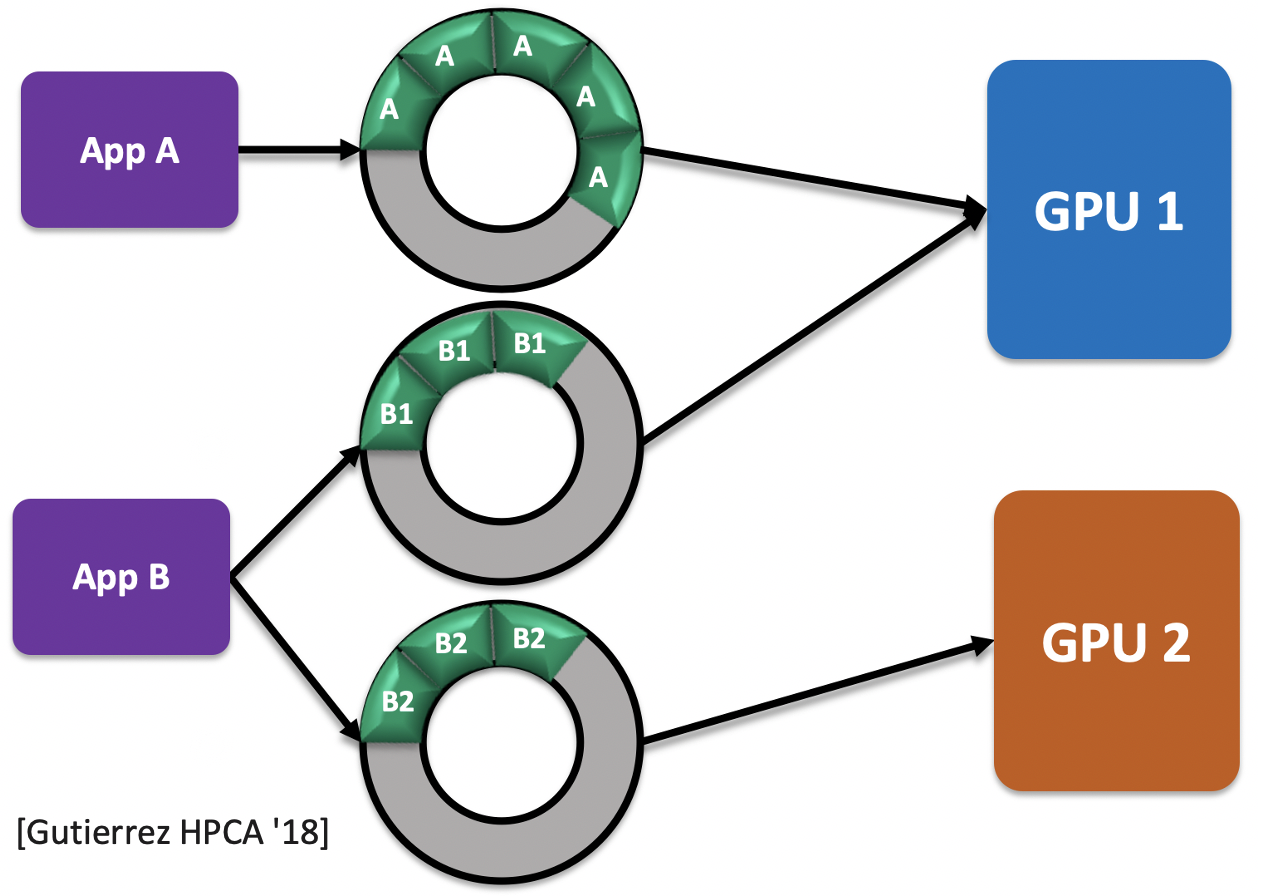
Once the GPU kernels/routines is converted to a task packet, a series of dependency analysis is executed on all the existing packets to identify the correct order of execution. Enqueuing packets maintains the order of execution into a task queue (implemented as a circular buffer in the above figure). If a task is entirely independent of other tasks, then it is enqueued to the top of the queue as it doesn't need to wait for other tasks to finish before its turn. On the other hand, tasks dependent on one and the other get enqueued in order of their dependency. This is further improved by providing multiple queues for different applications as a GPU most likely isn't just executing a single application at a time. Doing so provides a better resource utilization where the GPU is kept busy with active tasks at hand rather than spinning and idle waiting on existing tasks to complete from other connected GPUs.
This is just scratching the surface, and real-life software stacks are much more complicated. But my objective is for all of us to take a moment to appreciate the effort and software layers put into providing these programming models to make our lives as application scientists just a tad bit easier.